About the Swedish ASV portal
The portal of Swedish Amplicon Sequence Variants (ASVs) is an interface to sequence-based observations in the Swedish Biodiversity Data Infrastructure (SBDI). In addition to a database of amplicon sequence variants (ASVs) and a web interface, we provide a step-by-step guide(Link opens in a new tab) to submitting raw sequence data to ENA, and a pipeline(Link opens in a new tab) (developed in the nf-core collaboration) for denoising and taxonomic annotation of metabarcoding data.
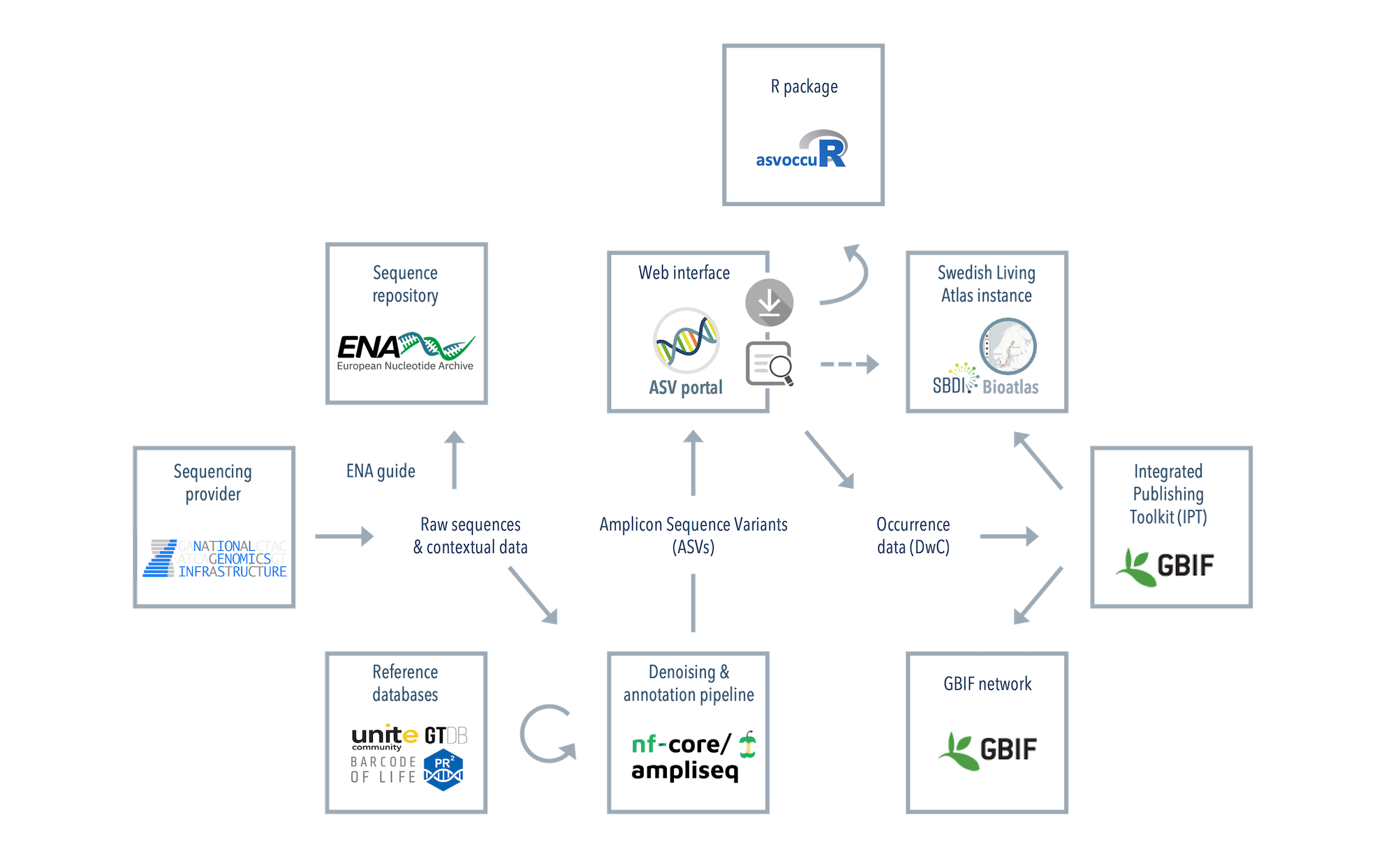
Flow of metabarcoding data in SBDI: A sequencing service provider, such as NGI, delivers sequencing data
to the user. The user submits raw sequencing data and contextual data to ENA
(see our guide(Link opens in a new tab)), denoises the data using e.g.
nf-core/ampliseq
(Link opens in a new tab)
, and submits the denoised data (ASVs and their counts in different samples, and contextual data)
to the ASV portal. Metabarcoding data in the ASV database/Bioatlas
can be searched and downloaded in a condensed format using the portal,
and downloaded archives can then be unpacked, merged and processed into ASV table format using the
asvoccur R package.
Why publish data in SBDI?
By publishing your data in the Swedish ASV database and Bioatlas your data will be easily reusable by others and can be seamlessly analysed in conjunction with other datasets in the Bioatlas. The ASVs will be regularly re-annoted as new reference database versions are released; thus your data will be updated with the latest taxonomic information. The ASV database also provides a complement to sequence repositories such as ENA or NCBI that typically only store raw sequencing data. Data in the Bioatlas is further regularly integrated in GBIF and therefor your metabarcoding-based biodiversity data will contribute to global knowledge about biodiversity.
Available data
Currently, we support inclusion of ASVs from the 16S rRNA gene for prokaryotes, the 18S gene for eukaryotes, the internal transcribed spacer (ITS) for fungi, and the COI cytochrome c oxidase I (COI) for metazoans. Other markers can be included on request. Read more about ASV taxonomy in SBDI here.
| Marker | Domains/Kingdoms/Supergroups* | # Datasets | # Phyla | # Classes | # Orders | # Families | # Genera | # Species | # ASV:s |
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA | Archaea, Bacteria, Unassigned | 3 | 120 | 305 | 966 | 2232 | 6639 | 12978 | 120064 |
| 18S rRNA | Amoebozoa, Archaeplastida, Archaeplastida:plas, CRuMs, Cryptista, Cryptista:nucl, Eukaryota_X, Excavata, Haptista, Obazoa, Provora, TSAR, TSAR:nucl, Unassigned | 4 | 32 | 207 | 443 | 891 | 2134 | 3194 | 51288 |
| COI | Animalia, Chromista, Plantae, Unassigned | 10 | 24 | 57 | 208 | 1179 | 6200 | 15803 | 892376 |
| ITS|LSU rRNA | Fungi | 4 | 13 | 43 | 99 | 216 | 346 | 616 | 1927 |
* To accommodate data from different reference databases, and to facilitate data transfer to the Bioatlas and GBIF platforms, we group GTDB domains and PR2 supergroups together with UNITE and BOLD BIN kingdoms here. Read more about ASV taxonomy in SBDI.
We are currently working on improving the accessibility of this website. Please contact SBDI support with any questions or suggestions!
